Prediction using rstanarm
postSurvfit.Rdposterior survival probability estimates from rstanarm for BIG data
Examples
# \donttest{
##
library(survival)
library(dplyr)
jmstan<-jmstanBig(dtlong=long2,
dtsurv = surv2,
longm=y~ x7+visit+(1|id),
survm=Surv(time,status)~x1+visit,
samplesize=200,
time_var='visit',id='id')
#> Fitting a univariate joint model.
#>
#> Please note the warmup may be much slower than later iterations!
#>
#> SAMPLING FOR MODEL 'jm' NOW (CHAIN 1).
#> Chain 1:
#> Chain 1: Gradient evaluation took 0.00084 seconds
#> Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 8.4 seconds.
#> Chain 1: Adjust your expectations accordingly!
#> Chain 1:
#> Chain 1:
#> Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
#> Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
#> Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
#> Chain 1:
#> Chain 1: Elapsed Time: 33.025 seconds (Warm-up)
#> Chain 1: 12.147 seconds (Sampling)
#> Chain 1: 45.172 seconds (Total)
#> Chain 1:
#> Fitting a univariate joint model.
#>
#> Please note the warmup may be much slower than later iterations!
#>
#> SAMPLING FOR MODEL 'jm' NOW (CHAIN 1).
#> Chain 1:
#> Chain 1: Gradient evaluation took 0.001033 seconds
#> Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 10.33 seconds.
#> Chain 1: Adjust your expectations accordingly!
#> Chain 1:
#> Chain 1:
#> Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
#> Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
#> Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
#> Chain 1:
#> Chain 1: Elapsed Time: 34.428 seconds (Warm-up)
#> Chain 1: 22.882 seconds (Sampling)
#> Chain 1: 57.31 seconds (Total)
#> Chain 1:
#> Fitting a univariate joint model.
#>
#> Please note the warmup may be much slower than later iterations!
#>
#> SAMPLING FOR MODEL 'jm' NOW (CHAIN 1).
#> Chain 1:
#> Chain 1: Gradient evaluation took 0.000795 seconds
#> Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 7.95 seconds.
#> Chain 1: Adjust your expectations accordingly!
#> Chain 1:
#> Chain 1:
#> Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
#> Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
#> Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
#> Chain 1:
#> Chain 1: Elapsed Time: 45.89 seconds (Warm-up)
#> Chain 1: 13.692 seconds (Sampling)
#> Chain 1: 59.582 seconds (Total)
#> Chain 1:
#> Fitting a univariate joint model.
#>
#> Please note the warmup may be much slower than later iterations!
#>
#> SAMPLING FOR MODEL 'jm' NOW (CHAIN 1).
#> Chain 1:
#> Chain 1: Gradient evaluation took 0.000943 seconds
#> Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 9.43 seconds.
#> Chain 1: Adjust your expectations accordingly!
#> Chain 1:
#> Chain 1:
#> Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
#> Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
#> Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
#> Chain 1:
#> Chain 1: Elapsed Time: 40.295 seconds (Warm-up)
#> Chain 1: 12.659 seconds (Sampling)
#> Chain 1: 52.954 seconds (Total)
#> Chain 1:
#> Fitting a univariate joint model.
#>
#> Please note the warmup may be much slower than later iterations!
#>
#> SAMPLING FOR MODEL 'jm' NOW (CHAIN 1).
#> Chain 1:
#> Chain 1: Gradient evaluation took 0.000843 seconds
#> Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 8.43 seconds.
#> Chain 1: Adjust your expectations accordingly!
#> Chain 1:
#> Chain 1:
#> Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
#> Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
#> Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
#> Chain 1:
#> Chain 1: Elapsed Time: 36.032 seconds (Warm-up)
#> Chain 1: 17.002 seconds (Sampling)
#> Chain 1: 53.034 seconds (Total)
#> Chain 1:
#> Warning: Bulk Effective Samples Size (ESS) is too low, indicating posterior means and medians may be unreliable.
#> Running the chains for more iterations may help. See
#> https://mc-stan.org/misc/warnings.html#bulk-ess
#> Warning: Tail Effective Samples Size (ESS) is too low, indicating posterior variances and tail quantiles may be unreliable.
#> Running the chains for more iterations may help. See
#> https://mc-stan.org/misc/warnings.html#tail-ess
mod1<-jmstan
P2<-postSurvfit(model<-mod1,ids<-c(1,2,210))
pp1<-plot(P2$p1[[1]])
pp1
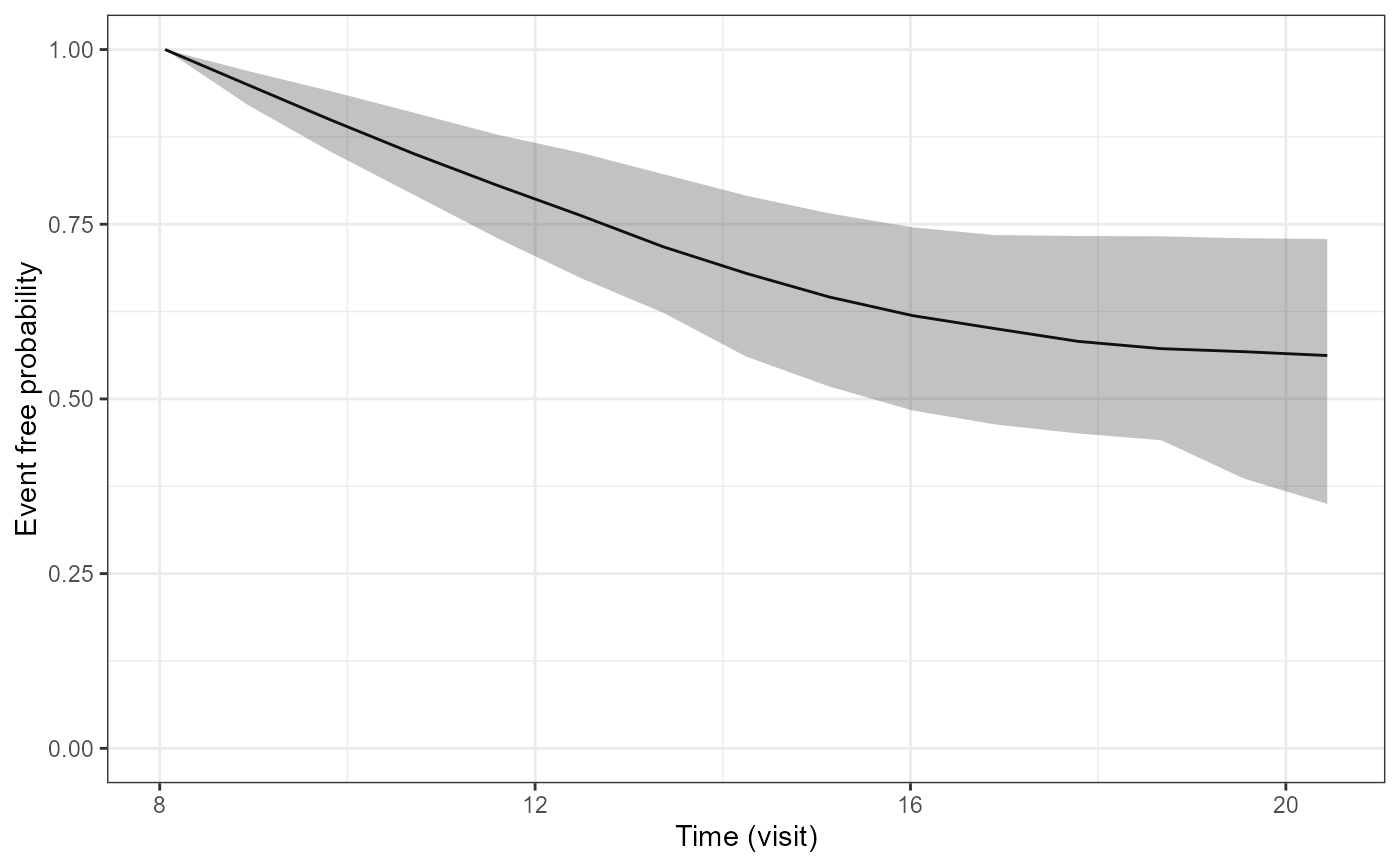 pp2<-plot(P2$p1[[2]])
pp2
pp2<-plot(P2$p1[[2]])
pp2
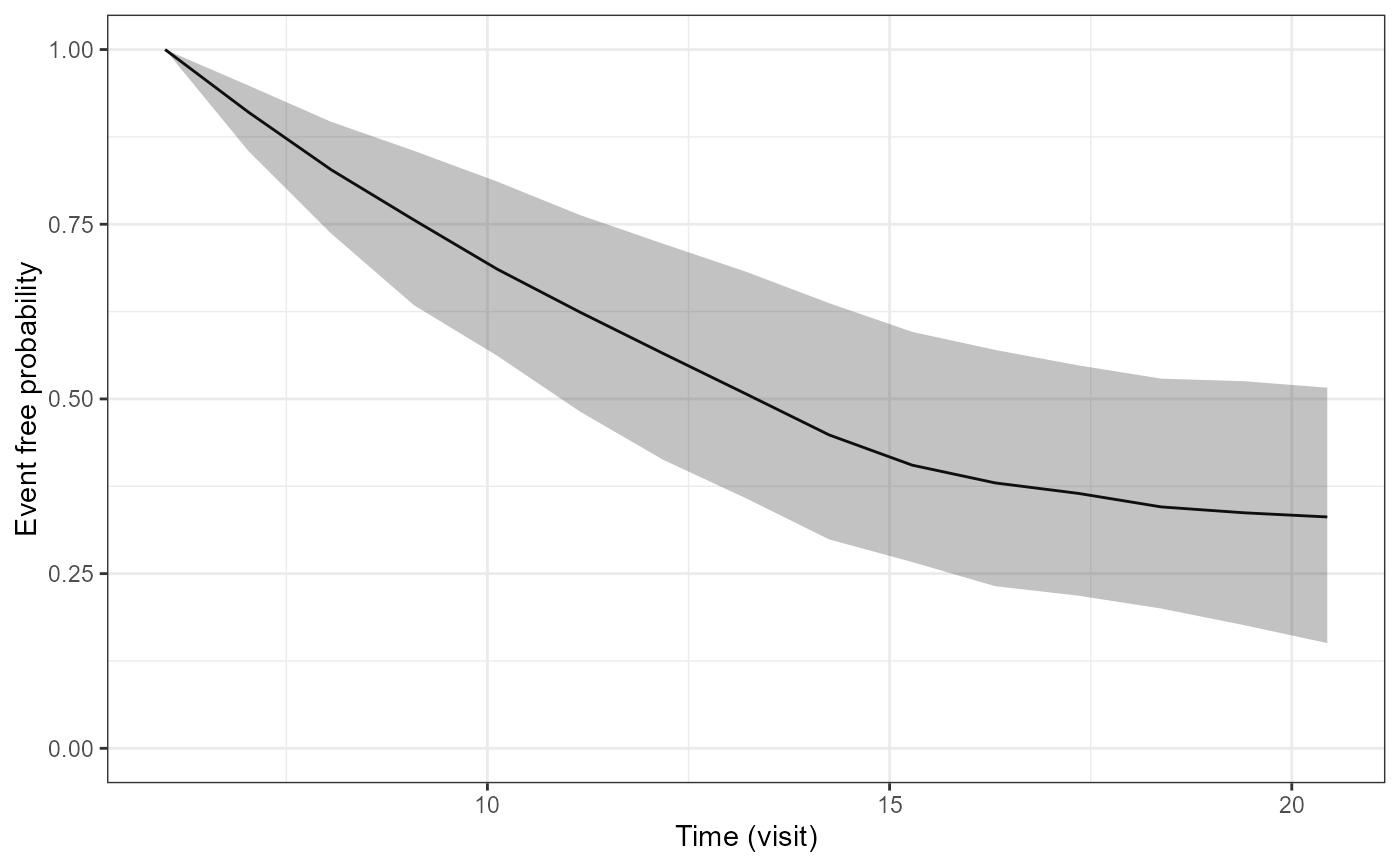 pp3<-plot(P2$p1[[3]])
pp3
pp3<-plot(P2$p1[[3]])
pp3
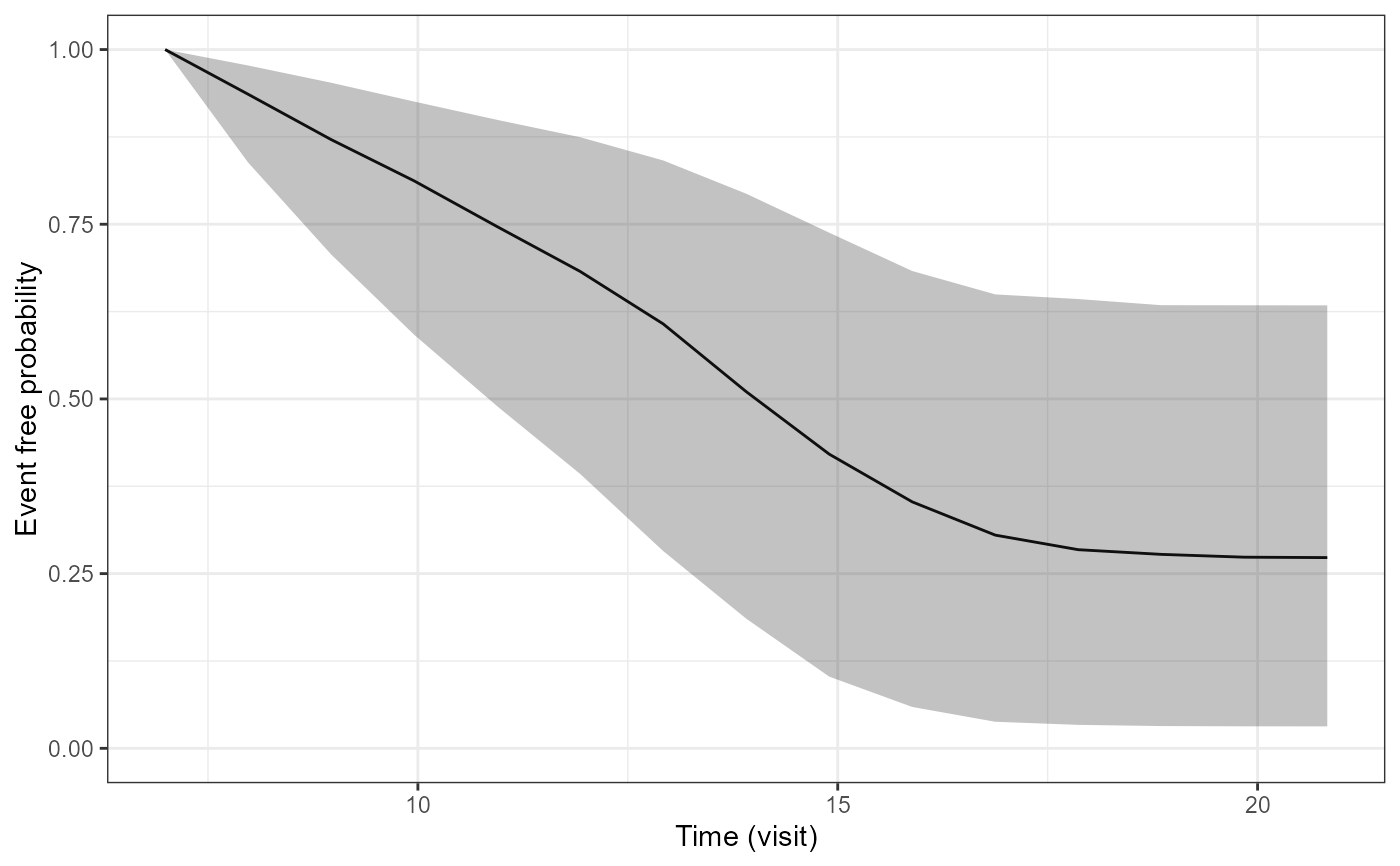 ##
# }
##
# }